Abstract Based ON The Crystal Structure of of Imatinib and tyrosine kinase Complex (the PDB ID: 2HYY)., The Active Structure and pharmacophores of Imatinib, and The Interaction BETWEEN Imatinib and The Active Site of tyrosine kinase have have been Explored It WAS found that The methylpiperazin Group In imatinib bound to the active site of tyrosine kinase weaker than other substructure, and there are no important pharmacophores, so this part structure of imatinib can be rebuilt. Further molecular design of anti-cancer drug imatinib’s analogues was carried out by De Novo Drug Design Method, 199 molecules were obtained at first, and 10 molecules with high score were screened by docking software, this research results shows that imatinib ‘s analogues with potential for effective tyrosine kinase inhibitors.
KeyWords anti-cancer drug; imatinib; analogue; De Novo drug design
Initio Molecular Design of Antitumor Drugs Imatinib Derivatives
Abstract In this paper, the structure of tyrosine kinase and imatinib complex crystals (PDB ID: 2HYY) , the active structure of imatinib, the pharmacophore and the mode of action of imatinib and tyrosine kinases were analyzed. Studies have shown that the piperazine moiety in the molecular structure of imatinib has a weaker effect on the enzyme, there is no important pharmacophore, and it is suitable for structural modification of this part of the structure; on this basis, the preliminary imatinib derivative was performed. De novo molecular design studies, designed 199 molecules, and used docking software to screen out 10 higher scoring molecules, indicating that imatinib derivatives are tyrosine kinase inhibitors. Key words anticancer drugs; imatinib; derivatives; de novo molecular design
1 Introduction
Novartis (Novartis) ‘s cancer drug Gleevec (Gleevec , Glivec) , the generic name imatinib mesylate (of Imatinib Mesylate) , code-named of STI571 , chemical name [4 – [(4- methyl yl -1- piperazinyl ) methyl ] -N- {4- methyl- 3- [4- (3 -pyridinyl ) -2 -pyrimidinyl amino ] – phenyl} – benzamide methanesulfonate . Its chemical structure is shown in Figure 1 .
FIG 1 imatinib of formula
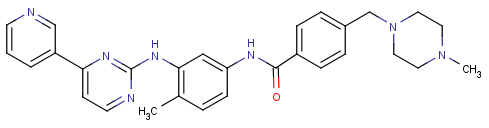
Fig.1 Structure of Imatinib
Chemistry Magazine gives you information about the Structure of Imatinib. Imatinib is a tyrosine kinase inhibitor drugs, 2001 years . 5 dated 10 May imatinib oral capsule formulation with anti-tumor mechanisms breakthrough into the US FDA express approval path for A – interferon to Drug failure Germ cell crisis crisis, chronic disease, accelerated disease treatment of granulocytic leukemia. The birth process of imatinib is another example of drug development [1] . The drug acts as an anticancer agent by “ turning off “ a tyrosine kinase that allows cells to have the ability to undergo canceration and proliferation.
Each year, there are approximately 4,500 chronic Myeloid Leukemia ( CML) patients worldwide and 23,000 diseased survivors. The last century 60 years, Nowell and Hungerford found CML patients with white blood cells contain an abnormal Philadelphia (Philadelphia) chromosome linked [2-3] . Then, 80 years,Witte et al. [4-5] found that this Philadelphia chromosome encodes a specific kinase gene, the BCR-ABL gene, which produces BCR-ABL kinase in the patient . This kinase catalyzes the phosphorylation of the substrate. Signals are transmitted within the cell to stimulate the proliferation of the patient’s myeloid cells, resulting in CML . In the early 1990s [6-8] , Zimermann et al. synthesized a series of small-molecule compounds mainly composed of 2 -phenylaminopyrimidines. After screening, it was found that CGP5714B , also known as STI571, can inhibit PDGFR tyrosine kinase in vitro [9] and later further demonstrated that this small molecule can specifically inhibit the activity of BCR-ABL [10] . Because of the excellent curative effect of imatinib, it was rated by Science as one of the top 10 technological breakthroughs in the world in 2001 [11] .
However, clinical studies in recent years have shown that imatinib has developed resistance to other tyrosine kinase inhibitors and has become a new research direction for anticancer drugs, in particular structural modifications and modifications to imatinib. In this paper, the active structures of imatinib, pharmacophores, and the mode of action of imatinib and tyrosine kinases were studied, and preliminary de novo molecular design of possible imatinib derivatives was attempted using computer-aided molecular design methods. Research and explore the feasibility of designing a novel tyrosine protein kinase inhibitor anticancer drug.
2 Research Methods The structure of the crystals of the tyrosine kinase and imatinib complexes(PDB ID:2HYY)was directly downloadedfrom the Protein Data Bank(PDB) [12] . First,LIGPLOTsoftware [13] was used to analyze the mode of action of imatinib and tyrosine kinases.Ligand Scout [14] software was used to analyze thepharmacophore information of imatinibin complex crystals(2HYY).LigBuildersoftware [15] andeHiTSsoftware [16] conducted de novo molecular design studies of tyrosine kinase inhibitors. The main research steps included the analysis of the three-dimensional structure of tyrosine kinase and imatinib complex crystals. ,Getinformation about tyrosine kinase active site; Then useGROWstrategy to generate a large number of candidate inhibitor molecules, and select the more excellent molecules; Finally, useeHiTSsoftware to dock these molecules with tyrosine kinase From this, choose the molecule with the lowest docking energy as the final research result. All calculations are done onAMD Althon 64 X2 3600+computers. All parameters use default values unless otherwise specified.
3 Results and discussion
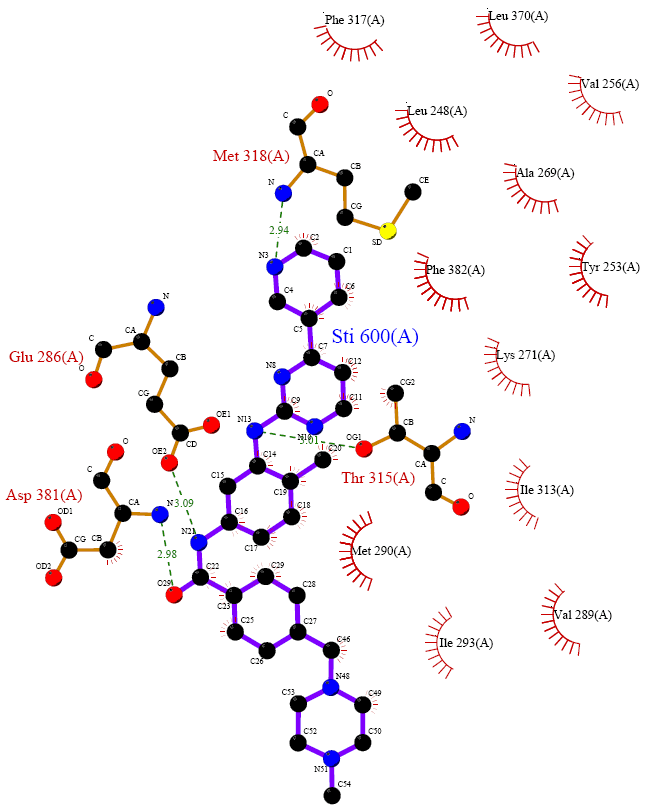
3.1 tyrosine kinase imatinib complex (2HYY) three-dimensional structures selected tyrosine kinase and the data structure of imatinib imatinib existing composite crystals (2HYY) as a three-dimensional structure of the original document, by the Figure 2 shows that the crystal structure is a tetrameric structure, gray spheres are four ligand molecules of tyrosine kinase, and four regions each represent an imatinib and tyrosine kinase complex. 3.2 tyrosine kinase with imatinib mode of action of FIG. 3 shows a Ligplot software depicted imatinib tyrosine kinase interacts with the structure of FIG. In the complex, the amino acid sequence of imatinib is STI 600. As can be seen in the figure, there are more amino acid fragments around imatinib in the complex of imatinib and tyrosine kinase, which is active. The sites are Phe317 , Leu370 , Leu248 , Val256 , Ala269 , Met318 , Phe382 , Tyr253 , Glu286 ,
Lys271 , Thr315 , Asp381 , Ile313 , Met290 , Ile293 , Val289 . Imatinib STI 600 and Met318 , Glu286 , Asp381 , 315 Thr there is a strong hydrogen bonding between the hydrophobic interaction between the fragment and the other belongs. Therefore, for the structure of imatinib itself, each functional group that generates hydrogen bonds should have a greater effect on the efficacy of the drug, while other structures that produce hydrophobic interactions have a smaller role, but for Drug molecules can act as skeletal support, so both have their own pharmacodynamic effects. More importantly, we have found that the piperazine ring of imatinib has almost no interaction with the enzyme. It was also reported in the literature that in the initial synthetic design of imatinib [17-18] , the 1 -methylpiperazine moiety in the molecule only increased the water solubility of the entire molecule, which was when the imatinib was bound to the enzyme. The structure is located outside the catalytic center of the enzyme, so the piperazine moiety does not affect the binding of imatinib to the enzyme. This finding can also be confirmed by the analysis of the following pharmacophores.
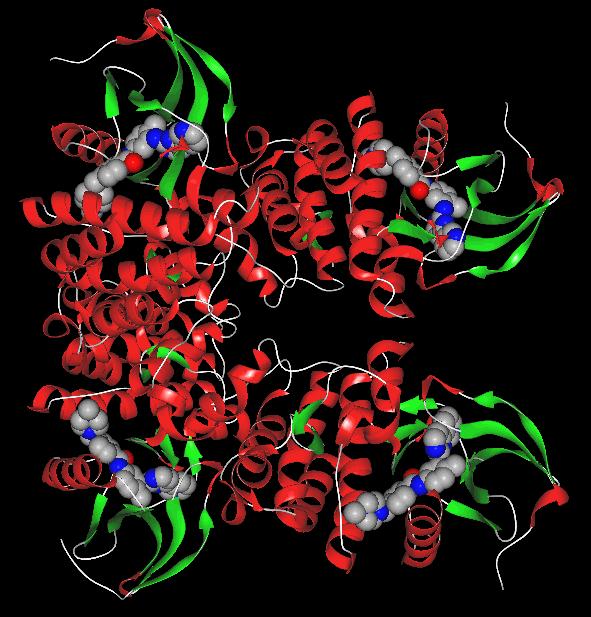
FIG 2 imatinib crystal structure of tyrosine kinase inhibitor and FIG nylon composite Fig.2 the Crystal Structure of Complex The Imatinib and tyrosine kinase
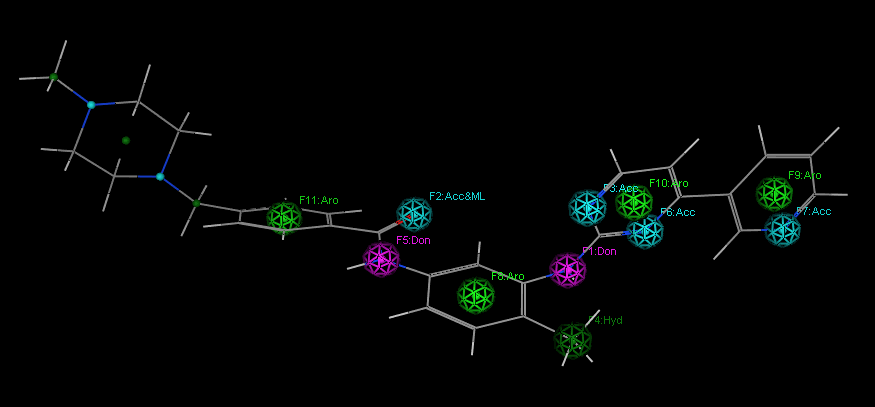
3.3 Pharmacophore analysis of imatinibFigure4gives information on the pharmacophore interacting with tyrosine kinase by imatinib. Among them,F1: DonandF5: Donrepresent a hydrogen bonding donor pharmacophore;F3: Acc,F6: AccandF7: Accrepresents a hydrogen bond receptor pharmacophore;F11: Aro,F8: Aro,F9: AroandF10: Arorepresents the central pharmacophore of the aromatic ring;F2: Acc&MLrepresents the hydrogen bond acceptor and metal coordination bond;F4: Hydrepresents the hydrophobic region. 3 imatinib tyrosine kinase interacts with a configuration diagram of Fig. 3 Structure of Imatinib bound to tyrosine kinase The Active Site of FIG. 4 by the imatinib interacts with tyrosine kinases resulting pharmacophore
Fig. 4 Pharmacophores of imatinib generated from imatinib and tyrosine kinase complex
From the pharmacophore analysis of imatinib, it can also be seen that the piperazine ring region does not have an important pharmacophore effect, and imatinib is suitable for structural modification of this part of the structure. We speculated that the piperazine ring was replaced by other groups. Afterwards, it may also inhibit the activity of tyrosine kinases. For this reason, we tried to perform preliminary de novo molecular design studies by using the remaining structure of the imatinib molecule as a template after removing the piperazine ring. It is expected that the activity may be relatively high or high. Active inhibitor molecule. If you have some questions or you want to learn more about chemistry in the news, feel free to contact us.
3.4 The molecular design process of imatinib
first uses the POCKET program to obtain information on the active pocket of tyrosine kinases. The program will generate key sites of action within the binding pocket. As shown in Figure 5 , the red region represents the hydrogen bond acceptor site, the blue region represents the hydrogen bond donor site, and the gray region represents the hydrophobic interaction site. The program produces two output files: one file stores all the molecules that make up the binding pocket; the other file stores all the grids in the binding pocket. These two files will be used in the subsequent ligand build program GROW .
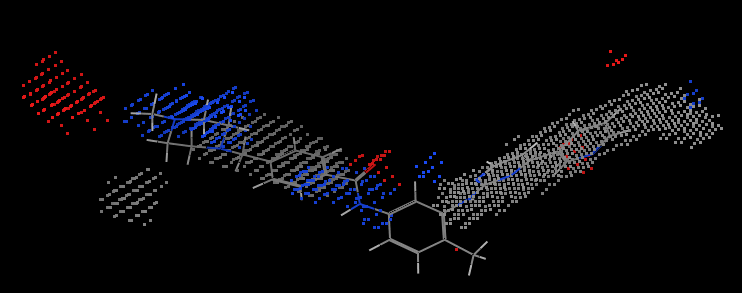
FIG . 5 2HYY key active site tyrosine kinase complex crystal structures of
Fig.. 5 of The Active Site of Key of Imatinib in 2HYY.pdb tyrosine kinase (Protein of The hidden for IS A Clear The representation of Sake. Shown of The ligand IS Here for instead to illustrate the shape of the binding pocket)
Throughout the analysis process, the source file 2HYY.pdb is first split into two files, the receptor tyrosine kinase .pdb and the ligand imatinib.pdb . Then the imatinib.pdb is converted to the imatinib.mol2 file using the format conversion program . the tyrosine kinase .pdb and imatinib.mol2 as input parameters programmed into pocket.index , running pOCKET module, to obtain the final file and the pharmacophore model described in the pocket receptor activity-related information.
Then use the GROW module to generate new molecules. Since it is the three-dimensional structural file 2HYY.pdb of the complex of tyrosine- labile ligand imatinib , it is desirable to isolate the “ seed “ structure from above imatinib . By analyzing the complex structure of tyrosine and imatinib, it can be found that the imatinib structure other than the piperazinyl group is a functional group of imatinib, so other parts were selected as the “ seeds “ for running the GROW module. structure. Firstly imatinib .pdb file to piperazinyl separated, saved as seed.pdbThe file is then converted into a mol2 format seed.mol2 file and hydrogenated. As the previous studies have shown that: piperazine ring region does not have important pharmacophore effect, this part of the structure is suitable for structural transformation, for this preliminary molecular design study for the original piperazine position of hydrogen as a “ seed ” ( Figure 6) structure of small ball position ) , to mark “ seed “ the growing point on, corresponding to the type of the hydrogen atom from “H” to “H.spc”.
6 FIG formula seed structure
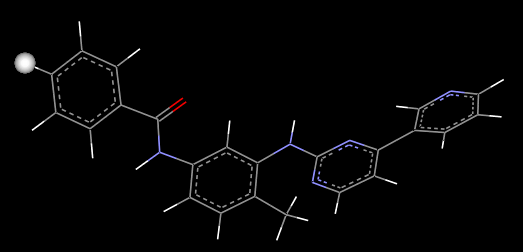
in Fig.6 Structure of Compound SEED
Finally deal with the new molecular library, run the PROCESS module to pick out the final result.
Using LigBuilder ‘s GROW module to construct a large number of ligand molecules, by analyzing the index files in each directory, comprehensive comparison of the molecular weight of each molecule, logP value, binding affinity and bioavailability, from which the value of logP was selected High molecular binding affinity and bioavailability, and smaller molecular weight molecules. The reason for this selection is: The molecular value of logP is high, and the molecular weight of the molecule is small, so it is highly soluble in lipids and easily penetrates the cell membrane and brain barrier, thereby facilitating the absorption. The molecules with strong binding affinity have strong interactions with the receptor protein and thus have strong inhibition; The use of strong molecules is easy to synthesize. According to these criteria, a total of 199 molecularly excellent ligand molecules were selected.
3.5 Docking Evaluation of Imatinib Derivatives
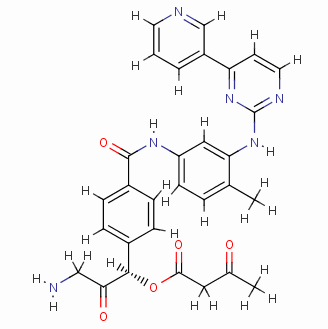
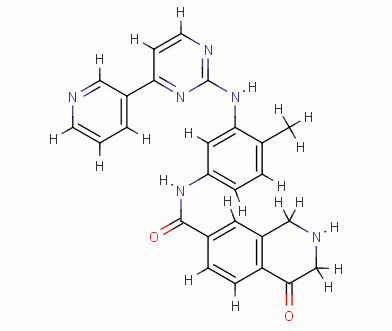
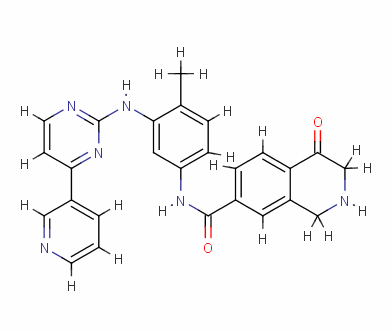
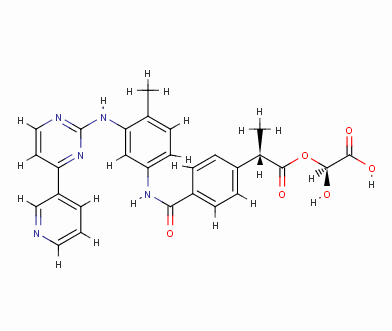
To reduce the amount of synthesis and experimental work in the future, the eHiTS software was used to further screen for excellent ligand molecules. The 199 molecules selected were docked one by one. Finally, the ten compounds with the highest scores were selected. The structure and docking energy of the ten high molecular molecules obtained are shown in FIG. 7 .
Result_085
eHiTS-Score: -10.5890
Result_002
eHiTS-Score: -10.3200
Result_133
eHiTS-Score: -9.7980
Result_043
eHiTS-Score: -9.7520
Result_095
eHiTS-Score: -9.5820
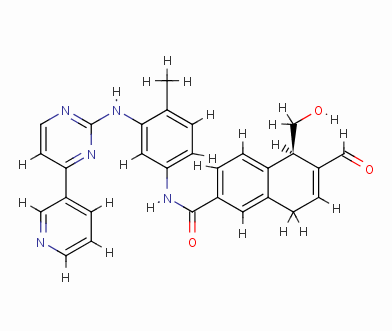
Result_098
eHiTS-Score: -9.3360
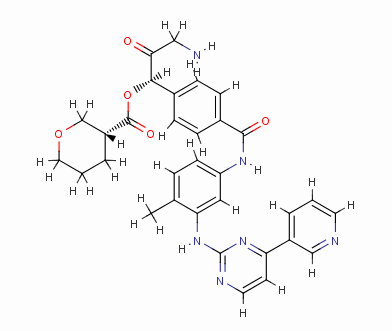
Result_003
eHiTS-Score: -9.1360
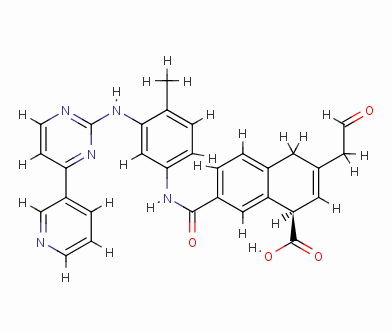
Result_074
eHiTS-Score: -8.8670
Figure 7 10 high-scoring imatinib derivatives
Fig.7 10 imatinib analogues with high score
From Figure 7 it can be seen that these designed high-grade imatinib derivatives have high scores, both greater than the imatinib score (eHiTS-Score: -7.2078) , theoretically better than imatinib . Higher activity. In addition, structural analysis of these 10 high-score imatinib derivatives revealed that the vast majority of these derivatives generate rings fused to the phenyl ring at the original piperazine ring and have carboxyl and hydroxyl groups on the ring. Polar groups such as carbonyl, and amino groups, on the one hand, in this part of the structure to generate a specific spatial three-dimensional structure and tyrosine kinase fit, on the other hand, by increasing the hydrogen-bonding, hydrophobic and other new binding interactions, it is possible Improving drug efficacy shows that they have the value of further experimental research. At the same time, preliminary molecular design studies in this article also showed that the new type of anti-cancer drugs with imatinib derivatives is feasible. The author will make further more comprehensive de novo molecule design and synthesis studies. In order to discover chemical structures that are better and easier to synthesize.
REFERENCES
[1] Wang Yuxi . Foreign Medicine , 2002, 23 (3): 147-148.
[2] Zhang Jingbo . Chinese Journal of Medicine , 2003, 5(5): 577-579.
[3] Duffaud F, Blay JY. Oncology, 2003, 65(3): 187-197.
[4] Witte ON, Dasgupta A, Baltimore D. Nature, 1980, 283: 826-31.
[5] Witte ON, Goff S, Rosenberg N, et Al. Proc Natl Acad Sci USA, 1980, 77: 4993-4997.
[6] Zimmermann J, Buchdunger E, Fabbro D, et al. Book of Abstracts, 212th ACS National Meeting, Orlando, PL, August 25-29, 1996 .
[. 7] Zimmermann J, G Caravatti, Mett H, et Al .. Archiv der Pharmazie, 1996, 329 (. 7):. 371-376
. [. 8] J. Zimmermann US 5,521,184, 1996-05-28
[. 9] Weisberg E, Griffin J D. Blood, 2000, 95: 34981.
[10] Druker BJ, Lydon N B. Clin Invest, 2000, 105(1): 3-7.
[11] The news and editorial staffs: Breakthrough of the year. Science, 2001, 294: 2443-2447.
[12] ] Cowan-Jacob SW, Fendrich G., Floersheimer A., et al. Acta Crystallorg., Sect. D, 2007, 63: 80-93.
[13] Wallace AC, Laskowski RA, Thornton J M. Prot Eng, 1995 , 8: 127-134.
[14] Wolber G, Langer T. J Chem Inf Model, 2005, 45(1): 160-169.
[15] Wang R, Gao Y, Lai L. J Mol Model, 2000, 6: 498-516.
[16] Zsolt Z, Darryl R, Aniko S, et al. Current Protein and Peptide Science, 2006, 7: 421-435.
[17] Zimmermann J, Furet P, Buchdunger E. In Anticancer Agents Frontiers in Cancer Chemotherapy. ACS Symposium Series 796.
[18] Ojima I, Vite G, Altmann K, et al. American Chemical Society: Washington, DC, 2001, 245-259
